About Glyco3d
Glyco3D features a family of databases covering the 3D features of monos, di, oligo, polysaccharides, glycosyltransferases, lectins, monoclonal antibodies and glycosaminoglycan-binding proteins. Other databases are also made available that completes the picture of glycan 3D structure decoding. that are made freely available to the scientific community. A search engine has been developed that scans the full content of all the data bases for queries related to sequential information of the carbohydrates or other related descriptors. This database ensemble offers a unique opportunity to characterize the 3D features that a given oligosaccharide can assume in different environments.
Jmol:
an open-source Java viewer for chemical structures in 3D is needed; it can It can be downloaded from SourceForge at this address:
http://jmol.sourceforge.net/
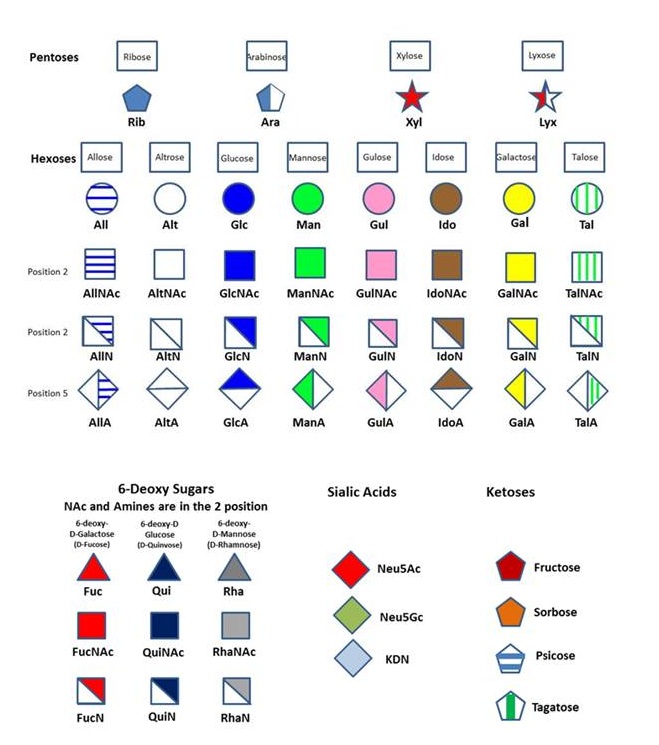
A common nomenclature has been adopted that conforms to the recommendations for carbohydrates and including the constraints required by the developing field of glycobiology in terms of visualization and encoding. The set of following rules specifies the nomenclature used throughout all the data bases.
Residue Letter Name: Rib, Ara, Xyl, Lyx, All, Alt, Glc, Man, Gul, Ido, Gal, Tal, ….and abbreviated trivial name.
- 6Ac for 6-O-acetyl group
- 3S for 3-O-sulfate group
- 6P for 6-O-phosphate group
- 6Me for 6-O-methyl group
- 36Anh for 3,6-anhydro
- Pyr for pyruvate group
Absolute Configuration: D or L The D-configuration for monosaccharide and the L configuration for Fucose and Idose are implicit and does not appear in the symbol. Otherwise the L configuration is indicated in the symbol, as in the case of Arabinose or L-Galactose.
Anomeric configuration: The nature of the glycosidic configuration (α or β) is explicitly set within the symbol.A detailed description of the monosaccharide nomenclature used in Glyco3D is presented in “The Symbolic Representation of Monosaccharides in the Age of Glycobiology” at www.glycopedia.eu
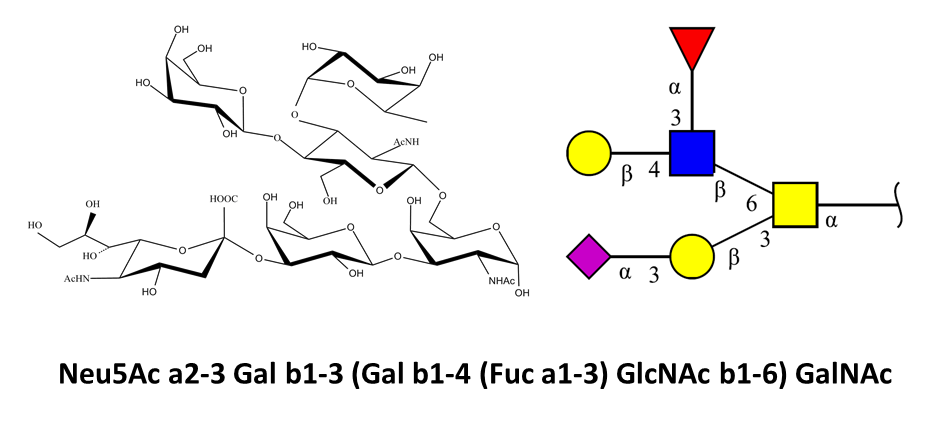
The syntax used to encode glycans in Glyco3D is exemplified below, along with some existing
notations and graphical representations.
Macromolecular builders are also made available for generating three-dimensional structures of polysaccharides, complex carbohydrates and complex solid state architectures such as glyco-gold nano particles.
Downloading Jmol. Jmol is an interactive web browser applet that is an open-source, cross-platform 3D Java visualizing tool for visualizing chemical and molecular structures. It provides 3D rendering with standard available hardware. . http://www.jmol.org/ Downloading the atomic coordinates for further independent and using a stand-alone viewer is another option
Contact us :
To contribute your structures to these data bases or for more information you can contact:
Centre de Recherches sur les Macromolécules Végétales (CERMAV-CNRS)
BP 53 X
38041 Grenoble cedex 9
France
Email : glyco3d@cermav.cnrs.fr
Phone : 33 (0) 76 03 76 36







